Yinglin Xia in Progress in Molecular Biology and Translational Science 2020. Unlike the 16S rRNA gene the ITS region is highly variable in length.

Ggtree For Microbiome Data R Bloggers Data Microbiome Data Analysis
The custom functions that read external data files and return an instance of the phyloseq-class are called importersValidity and coherency between data components are checked by the phyloseq-class constructor phyloseq which is invoked internally by the importers and is also the recommended function for creating a phyloseq object from manually imported data.
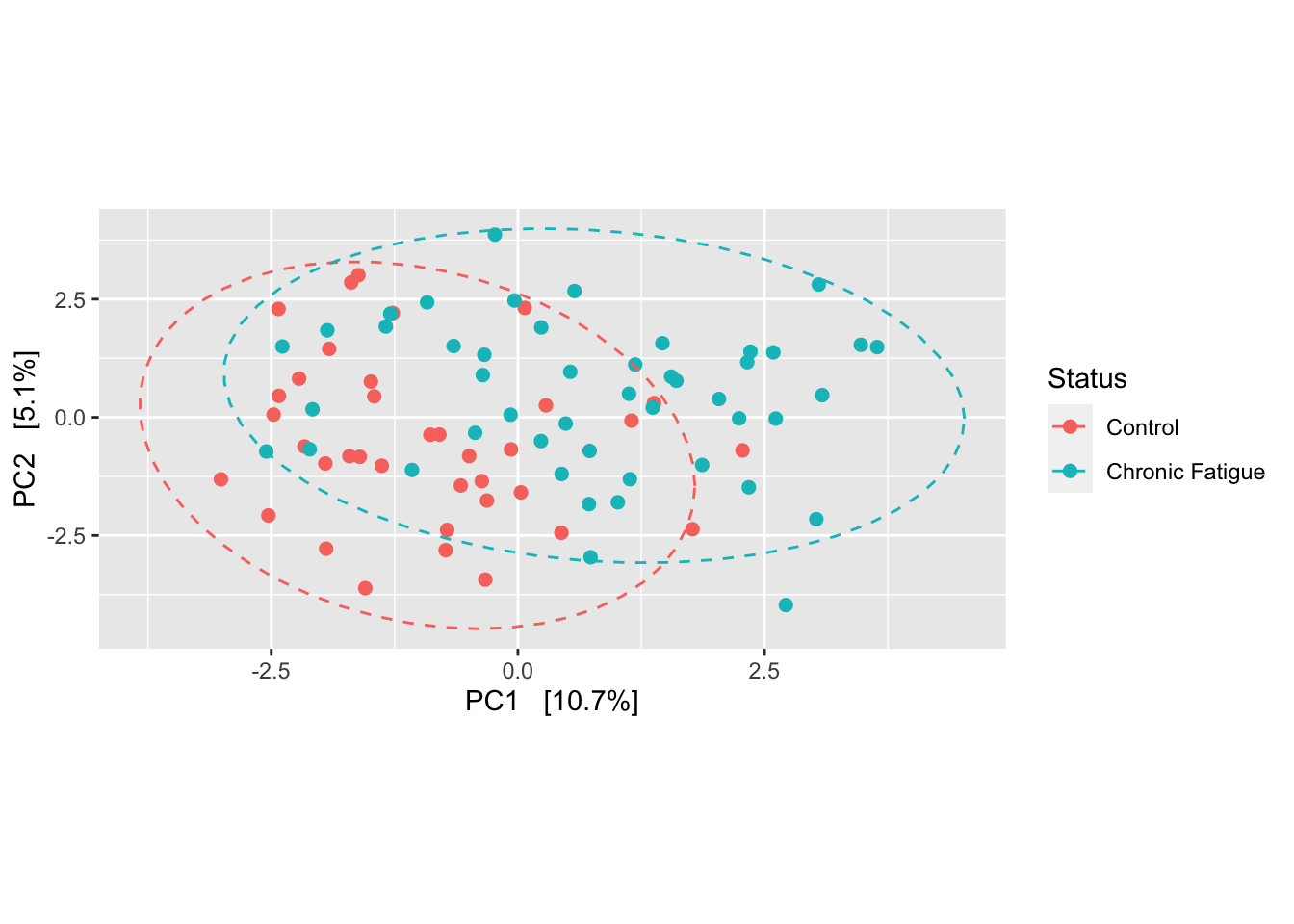
. The tutorial for R microeco file2meco and mecodev packages. An r Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. 17309896 Use cases Local comparison two of.
7132 Independent principal component analysis IPCA. Krumsiek J Arnold R Rattei T. In this paper we present a tutorial for scRNA-seq analysis and outline current best practices to lay a foundation for future analysis standardization.
The data are extremely noisy and large p and small n. We now demonstrate how to straightforwardly import the tables produced by the DADA2. The tutorial is using 2x250 V4 sequence data so the forward and reverse reads almost completely overlap and our trimming can be completely guided by the quality scores.
Cheetah Backronym for GEnome PAir - Rapid Dotter allows the calculation of dotplots even for large sequences like chromosomes or bacterial genomes. Zappia et al 2018. The key addition to this workflow compared to the tutorial is the identification and removal of primers from the reads and the verification of primer orientation and removal.
Compendium of 4941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Many tools can be used for the bioinformatic analysis of amplicon sequencing data such as QIIME Caporaso et al. IPCA 311 was proposed to solve the problems of both the high dimensionality of high-throughput data and noisy characteristics of biological data in omics studies.
Omics data have the problems. The commonly amplified ITS1 and ITS2 regions range from 200 - 600 bp in length. The challenges to standardization include the growing number of analysis methods 385 tools as of 7 March 2019 and exploding dataset sizes Angerer et al 2017.
The phyloseq R package is a powerful framework for further analysis of microbiome data. A rapid and sensitive tool for creating dotplots on genome scale.
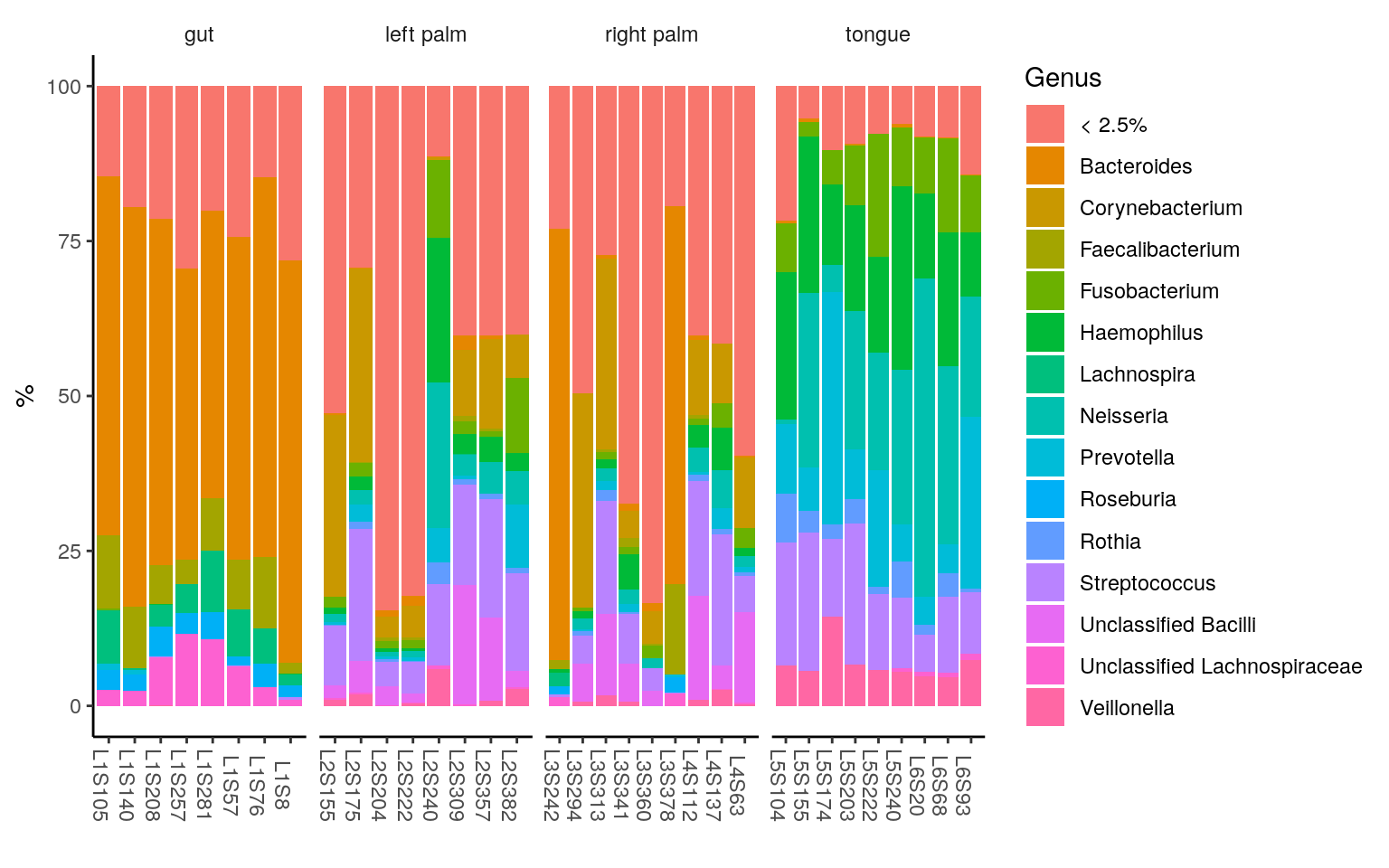
Tutorial For Microbiome Analysis In R Yan Hui
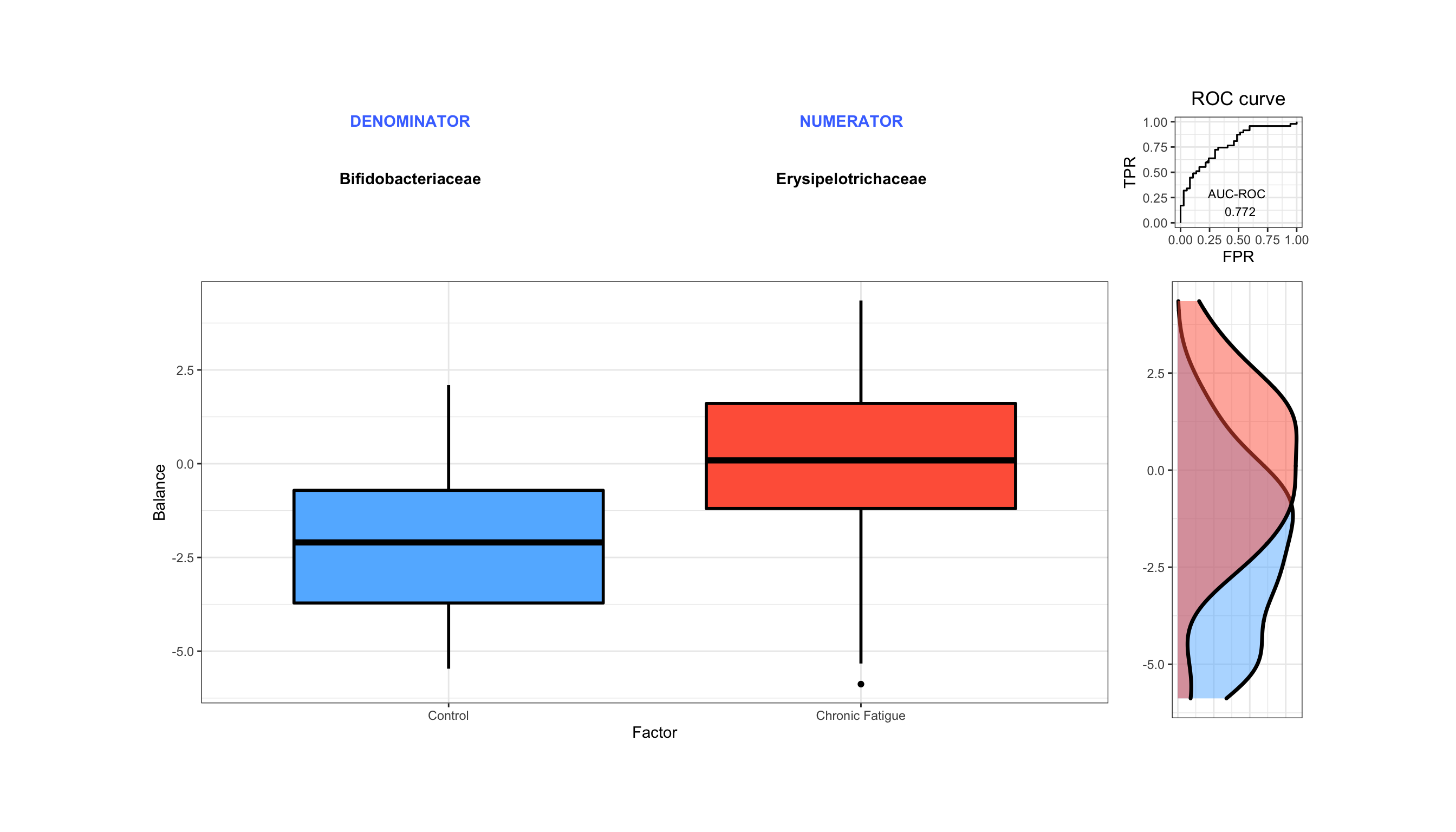
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
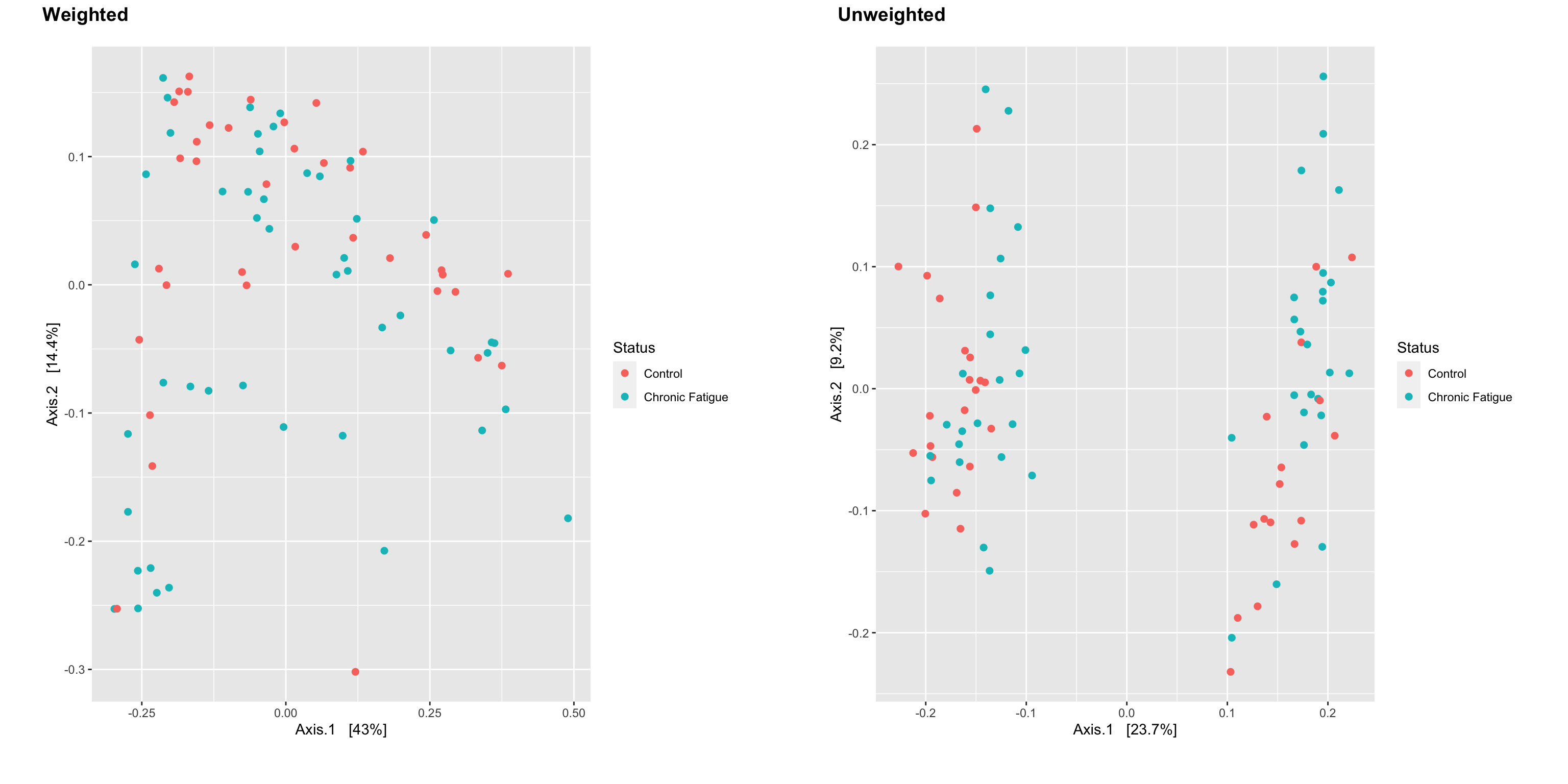
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
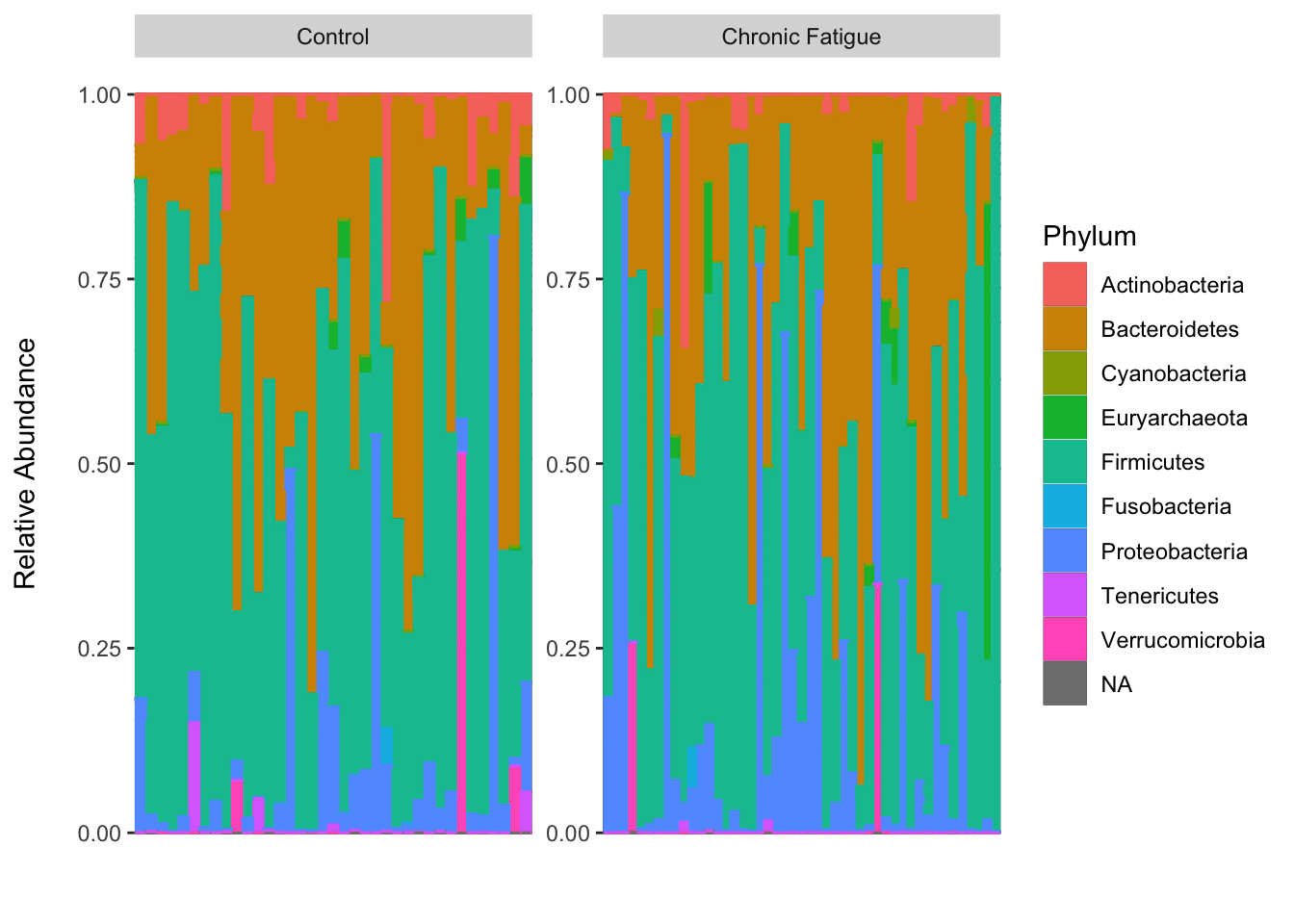
Introduction To The Statistical Analysis Of Microbiome Data In R Academic

Introduction To The Statistical Analysis Of Microbiome Data In R Academic
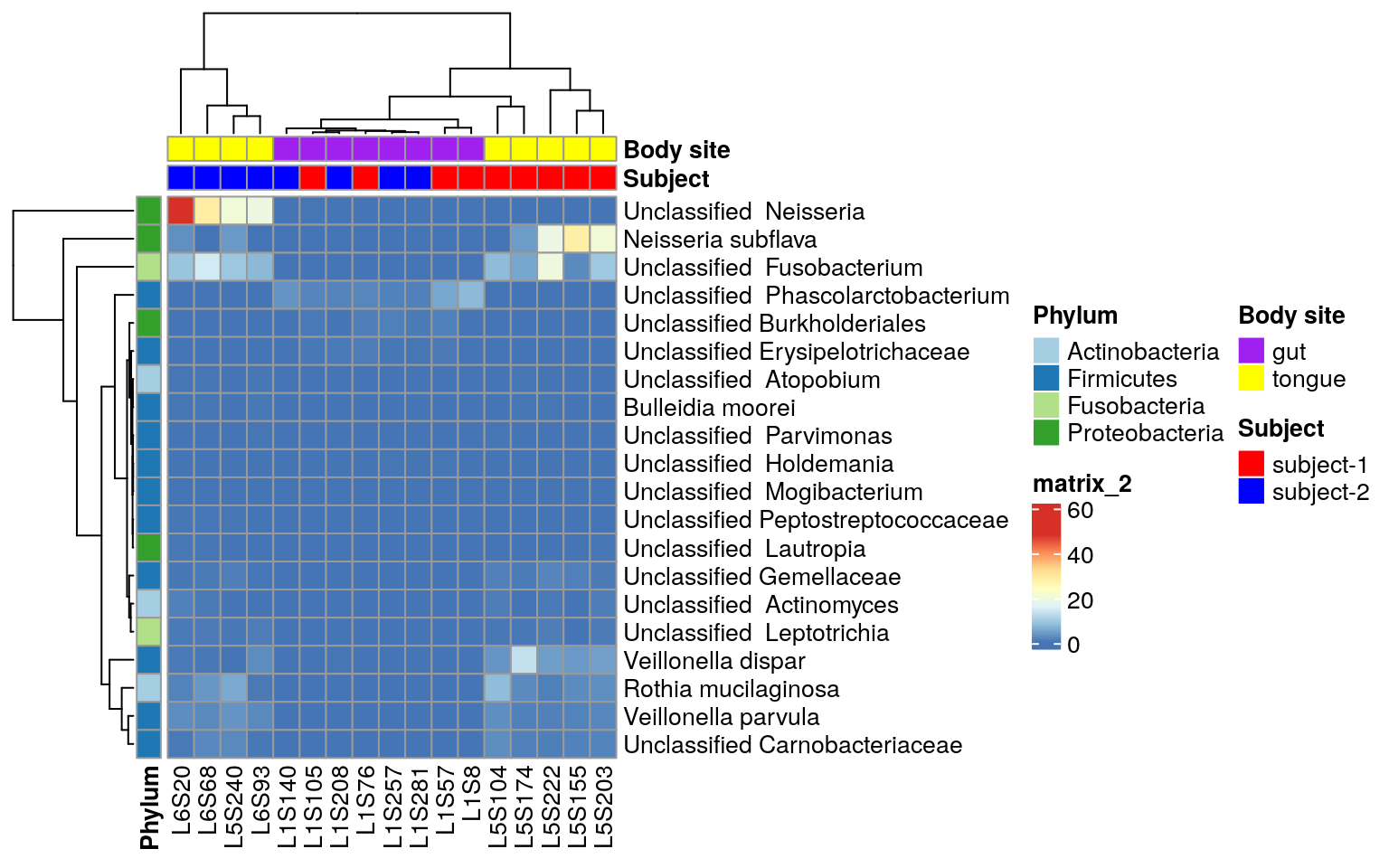
Tutorial For Microbiome Analysis In R Yan Hui
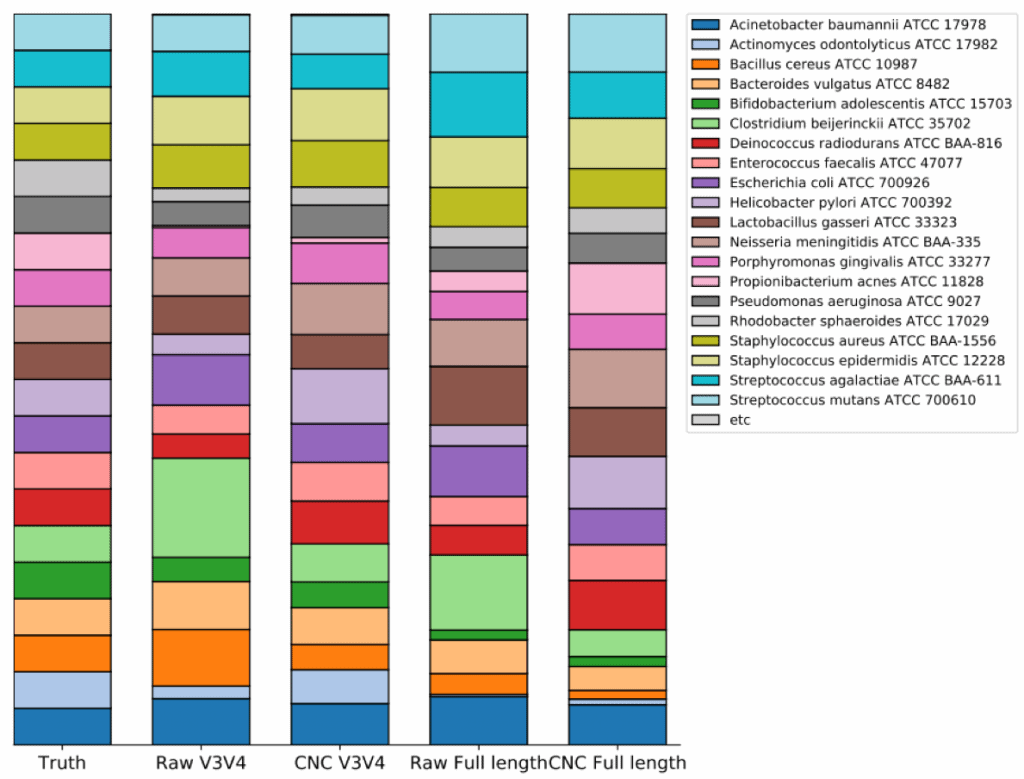
Tutorial 16s Microbiome Analysis Of Atcc R Microbiome Standards Ezbiocloud Help Center
0 comments
Post a Comment